Below is a listing of the academic projects I have been involved with. I have worked on projects involving the mathematical modelling of plants in studies of epidemiology and evolution, along with studies in bioinformatics and molecular biology.
You can find a list of my publications here.
Seed treatment differential equation based model (Rothamsted)
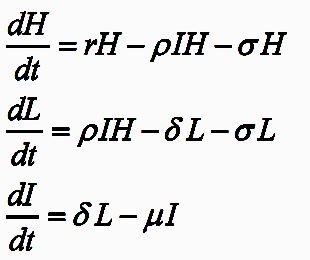
Read the paper here.
Fungicides can be applied as foliar sprays or seed treatments, but are susceptible to fungicide-resistance. To compare the selection pressures from both treatments I developed a differential equation based model in C++ that would calculate the fungicide effective life after the treatment types in isolation or in combination.
The model involved the simulation of the 11 leaf layers of
a winter wheat crop as infected by the pathogen Zymoseptoria tritici. Over time (measured as degree days) the volume of fungicide from the seed treatment pool would distribute amongst the leaf layers (imagine it like plumbing), and if specified at certain time points fungicide would be added as a spray.
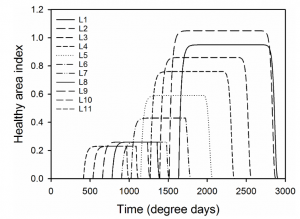
The model included a sensitive and resistant strain of pathogen, which would consume the plant healthy leaf area. Dose response curves would determine how much
the infected area of the sensitive strain would decline at certain fungicide concentrations, whereas the resistant strain would not decline at all, and so would be selected for over the simulated growing seasons. Once a certain proportion of healthy area was consumed, disease control was deemed lost, and so we would arrive at the effective life.
Plant Evolution Modelling: University of Warwick
The evolutionary processes behind crop domestication is what allowed us to cultivate plants and develop agriculture, making the shift from hunter-gathering to the reliance upon agriculture of today possible. Selection under domestication led to traits such as larger seed sizes and tougher grain rachises that have made plants more amenable to farming, yet the process of domestication is still unclear.
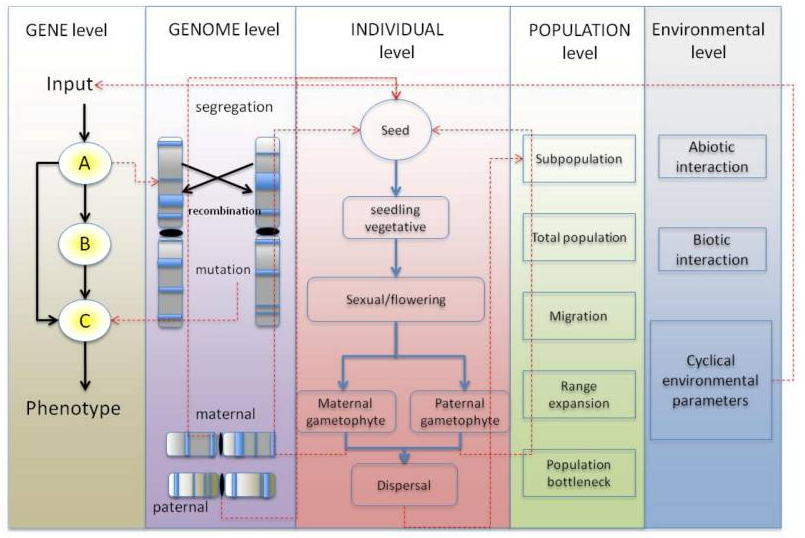
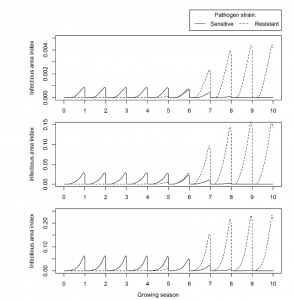
At the University of Warwick I developed a spatial agent-based model called Agrisims that was used to simulate the process using the Java programming language. Each simulated individual contained a collection of genes that were able to function in a network, take in an input from the environment and generate an output contributing to an individual’s trait and thus its fitness (for example, the degree of reliance on moisture, right). Mutation of the gene would thus impact on it’s fitness, leading to selection for certain alleles over others.
Elements of the work were published in peer review papers and at the 2012 conference for the Society of Molecular Evolution (SMBE). In the latter publication I used Approximate Bayesian Computation (ABC) to calculate liklihood densities of model parameters involving the tough grain rachis trait using observed archaeological data. Results showed that crops may have been harvested early in the year (green harvesting) lowering the selection pressure for this trait.

Cheap PCR primer design at the University of Warwick
Read the paper here
To be continued!